Our group at Case Western Reserve University combines experiments with theory and simulation to understand polymer physics problems involving grafted and non-linear polymers.
Computational techniques that we use include dissipative particle dynamics (DPD) simulations, polymer field theories such as self-consistent field theory (SCFT), and Monte Carlo simulations. Experimentally, we employ a wide range of techniques to investigate our polymer systems, including resistive pulse sensing (RPS), dynamic light scattering (DLS), depolarized DLS (DDLS), massively-parallel phase analysis light scattering (MP-PALS), multi-angle (static) light scattering (MALS), and a variety of elastic and inelastic neutron scattering techniques.
A SELECTION OF RECENT RESEARCH PROJECTS
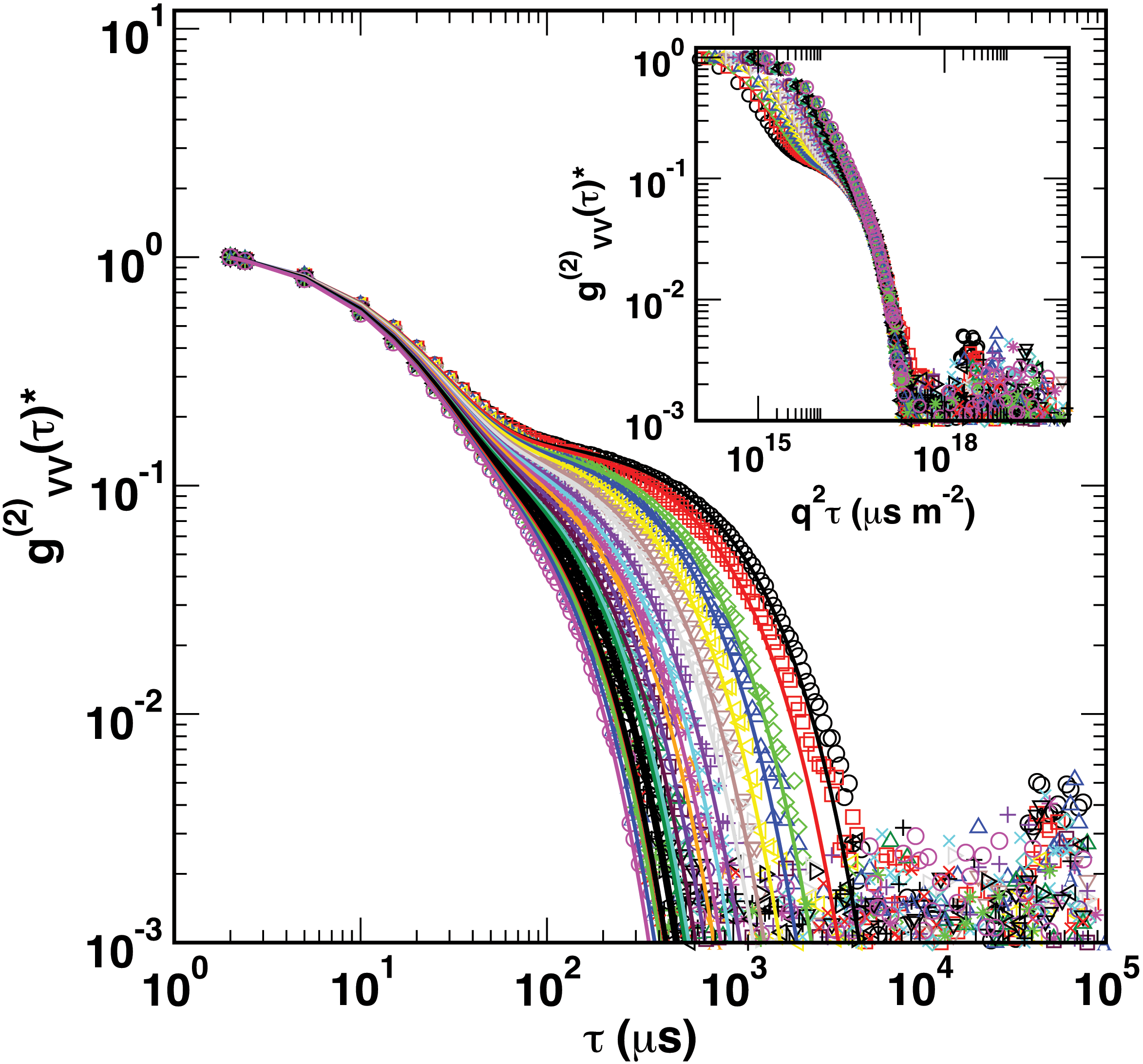
Depolarized DLS measurements of Au nanorods. Curves currespond to increasing values of the scattering vector q. The inset shows collapse of a portion of the curves, separating purely translational motion from rotational motion.
DEPOLARIZED DLS + GENETIC OPTIMIZATION
While small-angle scattering techniques — such as SAXS and SANS — are powerful methods to measure polymer brush structure on nanorod surfaces, its application to anisotropic nanoparticles can be challenging.
Depolarized dynamic light scattering (DLS) offers a fast and convenient way to measure translational and rotational diffusion coefficients of anisotropic particles. When plasmonic nanoparticles, such as Au nanorods, are used, wavelength-dependent depolarization ratios can lead to enhancements in light scattering intensities, further improving the capabilities of the technique. We apply a genetic optimization algorithm to optimize values of length and diameter of the nanoparticles in order to best match experimental relaxation rates – providing a technique that is complementary to our neutron scattering measurements.
Simulation of a single polymer-grafted nanorod using the rigid-body dynamics features of gDPD. Click to play video!
RIGID-BODY DPD SIMULATIONS
Our GPU-accelerated DPD software (gDPD) is now capable of performing full rigid-body dynamics simulations of nanoparticles, including those grafted with polymers.
The video on the left shows a simulation of a nanorod (aspect ratio 10) that is grafted with polymers (chain length 20). The simulation was performed for a million time steps, taking approximately 3 hrs to complete on an NVIDIA A-100 GPU.
Simulation of a polymer coil transitioning to a globule as the temperature increases above the critical temperature of the system, and unfavorable interactions kick in. Click to play video!
MACHINE-LEARNING + SIMULATIONS
Using gDPD, Proper Orthogonal Decomposition (POD), and Uniform Manifold Approximation and Projection (UMAP), we used DPD simulations to observe the relaxation modes of thermoresponsive polymers that undergo a coil-to-globule transition.
The dynamics of polymer chains were compared to single-chain nanoparticles, made by crosslinking a chain with itself. Read the full paper in Soft Matter. This paper was featured in the Soft Matter “Pioneering Investigators” special issue.
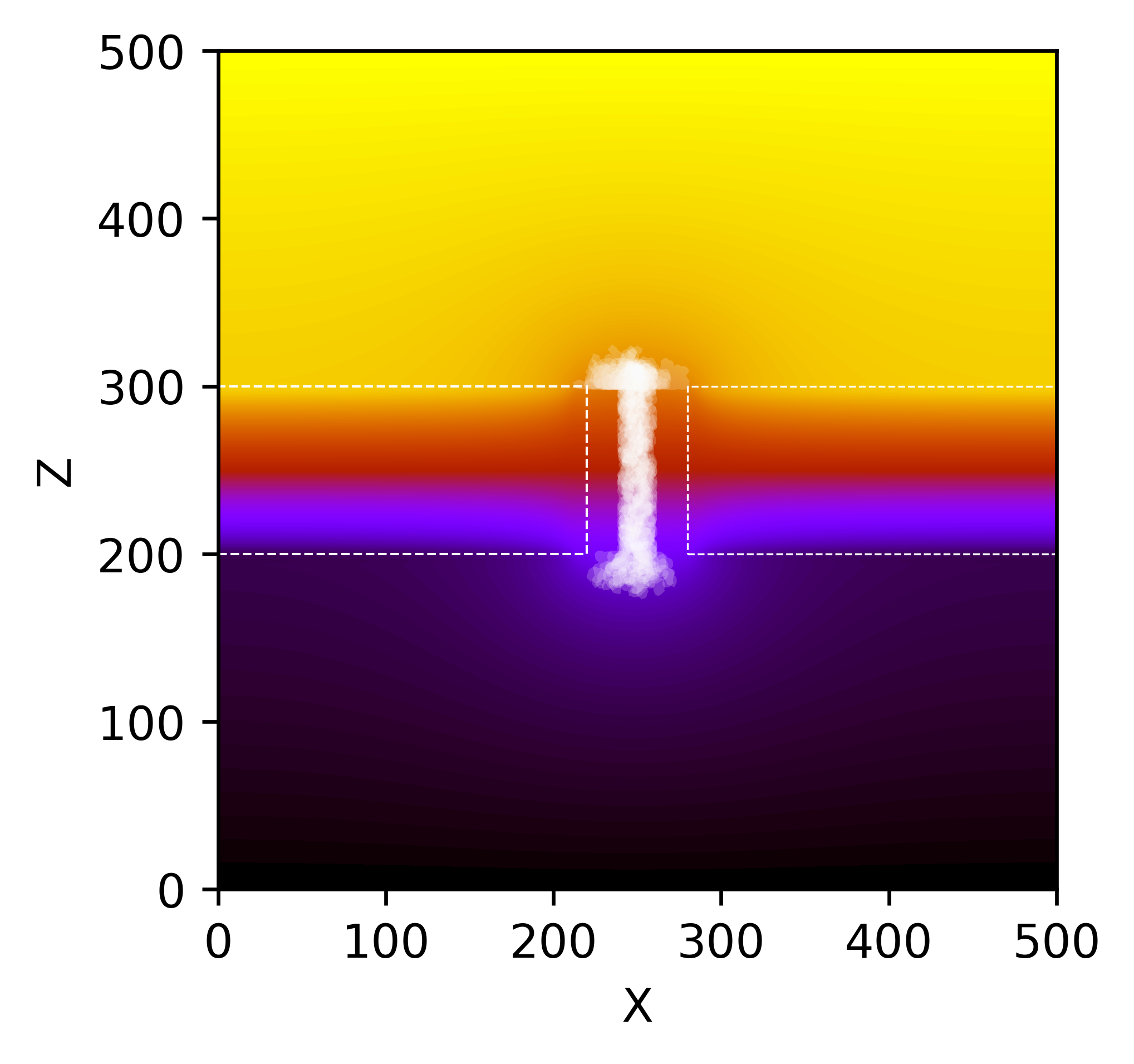
A plot of thousands of individual translocation events simulated using Dynamic Monte Carlo. For each position in the trajectories, the ionic current through the nanochannel was computed using the Poisson-Nernst-Planck (PNP) framework. PNP calculations were significantly accelerated by using a neural network.
MACHINE-LEARNING + RESISTIVE PULSE SENSING
Resistive pulse sensing (RPS) measurements of nanoparticle movement through individual nanopores are challenging experimental measurements to make, and their interpretation can be even more challenging! In this work, we combined experimental measurements with dynamic Monte Carlo simulations to study the behavior of large numbers of translocation events. We used the Poisson-Nernst-Planck framework to simulate the experimental translocation signatures. These calculations can be slow and must be done millions of times for a dynamic Monte Carlo simulation. We used a neural network to speed up the calculations of the ionic current through the nanochannel.
Read the full paper in Soft Matter!
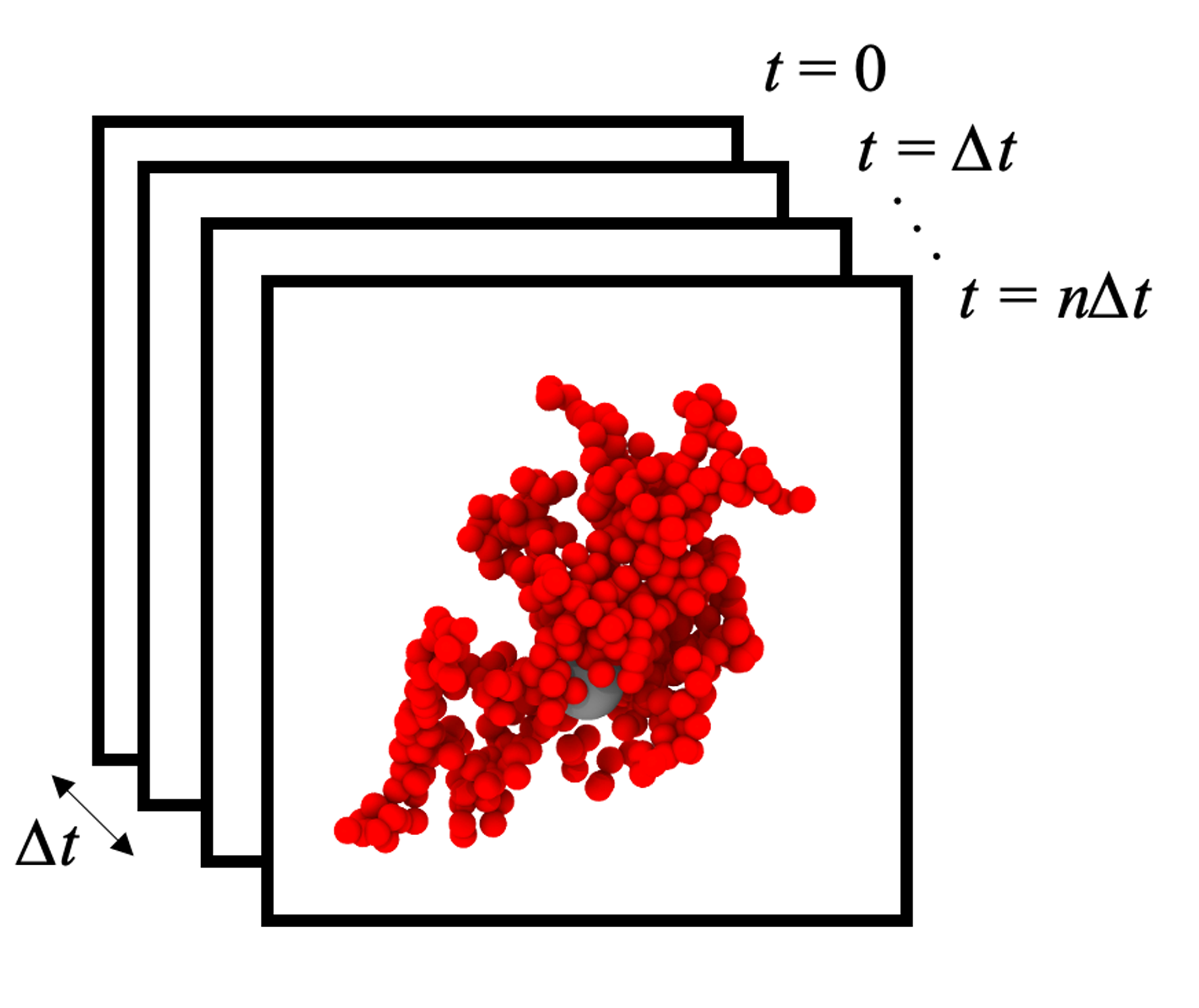
Depiction of the POD process. Snapshots are collected at regular intervals during the simulation, and post-processed to extract the relaxation modes of the particles and/or polymers. Click to play video!
DATA-DRIVEN ANALYSIS OF POLYMER DYNAMICS
Rouse Mode Analysis has been very successful for interpreting polymer relaxation dynamics from coarse-grained molecular dynamics simulations. However, it can be difficult to apply the technique to grafted polymers and non-linear architectures due to the fact that the relaxation modes depend very strongly on boundary conditions.
To overcome this limitation, we use a data-driven approach to extract the relaxation modes from the simulation trajectories using Proper Orthogonal Decomposition (POD) / Principal Component Analysis (PCA). These modes allow us to then compute relaxation times and interpret the dynamics of more sophisticated polymer architectures.
We have so far applied this to polymer-grafted nanoparticles (ACS Polymers Au) and molecular bottlebrushes (Macromolecules).